MaxCut Supertree
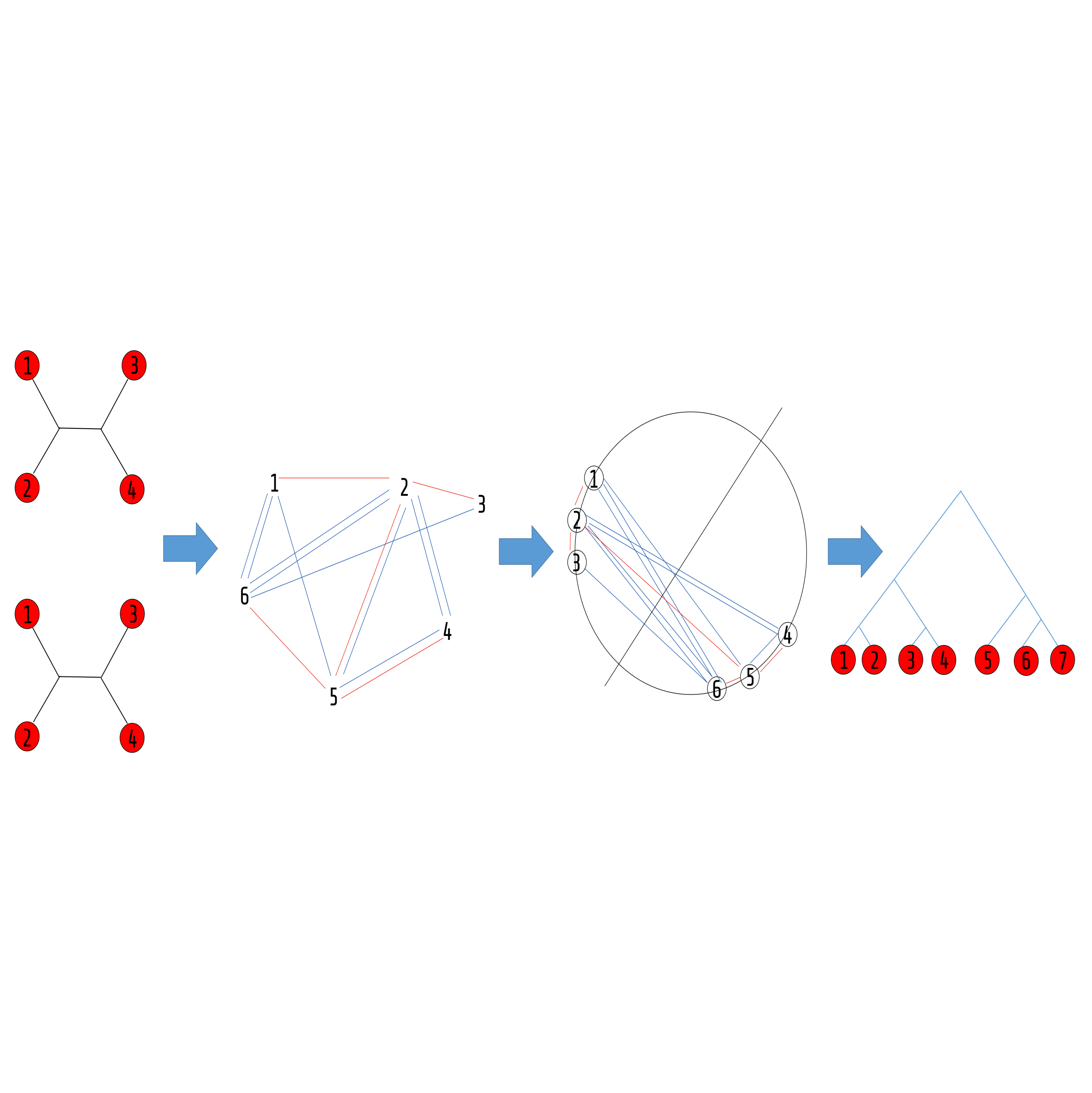
A quartet tree (or simply a quartet) is a tree over four taxa (species) that is the most basic phylogenetic building block – A tree can be decomposed into its defining quartets that uniquely define it. It is this fundamentality that confers the quartets their power. The converse however, is not true – it is rare that an arbitrary quartet set agrees on some tree. Hence, given a set of quartets, it is a common practice to look for a tree agreeing with the most of them. This seemingly simple and “clean” question lies at the heart of almost any specific, more complicated question in phylogenetics. Due to its centrality and importance, it has gained a lot of attention, from both theoreticians and practitioners. Notwithstanding, very little has been achieved since it was first introduced, more than two decades ago [1] – the best rigorous approximation to it, is obtained naively by a random tree, satisfying a third of the quartet set [2], and practical software use crude heuristics and are either slow or inaccurate.
During the years, our lab has dealt with many aspects of the supertree task, for the centrality and challenge of the associated tasks. Starting from my postdoc at Berkeley, we have developed a novel approach that embed the data in a graph and applies a MaxCut algorithm to the graph in order to classify the species according to the quartet information. The MaxCut, a very hard computational problem, is “solved” using a randomized semi-definite programming (SDP) approach that yields a very fast implementation. The general approach, quartet MaxCut has provided both theoretical and practical improvements in this well studied field that have been published in top conferences and journals in and bioinformatics [3,4,5] and computer science [6,7,8]. The approach (and software) is generic and was found useful in other applications such as classifying shapes in computer graphics [9], music performance analysis [10]. We were awarded the Israeli Science Foundation grant for this research direction. It is a nice example to the interrelationships between theory and practice in the field.

M. Steel. The complexity of reconstructing trees from qualitative characters and sub- tress. Journal of Classification, 9(1):91–116, 1992.
L. Gasieniec, J. Jansson, A. Lingas, and A. Ostlin. On the complexity of comput- ing evolutionary trees. In Proceedings of the 3rd Annual International Computing and Combinatorics Conference, pages 134–145, 1997.
S. Snir and S. Rao. Quartets MaxCut: A Divide and Conquer Quartets Algorithm.IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB), Volume 7(4): 704-718 (2010).
S. Snir and S. Rao. Quartet MaxCut: A Fast Algorithm for Amalgamating Quartet Trees.Molecular Phylogenetics and Evolution (MPE), 2012.
E. Avni*, R. Cohen*, and S. Snir. Weighted Quartets Phylogenetics. Systematic Biology. 64 (2): 233-242, 2015. *Authors contributed equally.
N. Alon, S. Snir and R. Yuster. On the compatibility of quartet trees, SIAM J. Discrete Math (SIDMA) . 28(3), 2014. Preliminary version appeared ACM-SIAM Symposium on Discrete Algorithms (SODA) 2014.
S. Snir and R. Yuster. Reconstructing approximate phylogenetic trees from quartet samples. SIAM Journal on Computing (SICOMP). 41(6): 1466-1480 (2012). Preliminary version appeared in SODA 2010.
S. Snir and R. Yuster. A Linear Time Approximation Scheme for Maximum Quartet Consistency on Sparse Sampled Inputs. SIAM J. Discrete Math. 25(4): 1722-1736, 2011. A preliminary version appeared in APPROX 2011
Shi-Sheng Huang, Ariel Shamir, Chao-Hui Shen, Hao Zhang, Alla Sheffer, Shi-Min Hu, and Daniel Cohen-Or. Qualitative organization of collections of shapes via quartet analysis. ACM Trans. Graph., 32(4):71:1–71:10, July 2013.
B. Chor E. Liebman, E. Ornoy. A phylogenetic approach to music performance analysis. Journal of New Music Research, 41(2):195–222, 2012.