!!!דרושים סטודנטים
New: Excellent Students are Needed!!!
Due to grants winning in joint projects with leading labs abroad, highly qualified students are needed. More details.
Research Areas:
- Phylogenetics
- Genomics
- Graph Theory
- Combinatorial Optimization
News:
From the Tiniest Creature,
to the Biggest Mammal:
Evolution, Learning, Bioinformatics
December, 14, 2023 From 14:00-18:00
Sfadia auditorium, Multi purpose Building
The seminar will be given by Mrs Yardena Peres from the IBM Watson Health “Analytics in the Real World: Predicting Hypoglycemia Events for People with Diabetes”.
Our Research
 Our lab focuses on mathematical and algorithmic approaches to bioinformatics problems in the fields of Evolution and Genomics. The problems we investigate are characterized by a scarcity of accurate modeling, or reliable, exact, and fast methods. This is partly due to the following reasons:
Our lab focuses on mathematical and algorithmic approaches to bioinformatics problems in the fields of Evolution and Genomics. The problems we investigate are characterized by a scarcity of accurate modeling, or reliable, exact, and fast methods. This is partly due to the following reasons:
Our lack of understanding of the biological factors influencing the evolutionary process.
Almost all problems arise in this field are computationally hard to solve.
We have sought to address these two issues by pursuing a number of directions:
Understand existing mathematical models and inspect how well they represent the actual biological processes.
Introduce new efficient algorithmic procedures to address established problems.
Develop new models for evolutionary processes using large biological data sets.
For our research, we use tools from fields such as combinatorial optimization, statistics and probability, mathematics, and information theory. Our research is characterized by ample collaborations with researchers from broad disciplines under the general framework of a systematic analysis of evolutionary processes aiming at finding biologically significant patterns.
Publications
Tracing Prokaryotic Evolution via Ordered Orthology
Tracing Prokaryotic Evolution via Ordered Orthology It is generally believed that the exponentially accumulating genetic molecular data will bring us closer to resolving one of
Horizontal Gene Transfer
Horizontal Gene Transfer Horizontal gene transfer (HGT), the passage of genetic material between genetically distant organisms, is a significant factor in microbial evolution. HGT plays
Publications
PUBLICATIONS Book Chapters J. Al-Aidroos and S. Snir, “Analysis of Point Mutations in Vertebrate Genomes”, in Algebraic Statistics for Computational Biology . Edited by L. Pachter
Research Projects
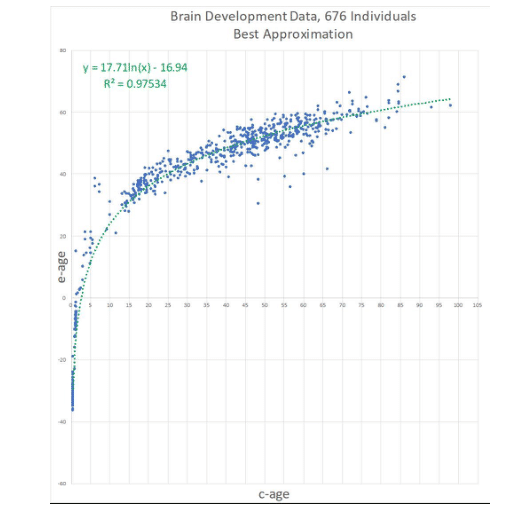
Epigenetic PaceMaker
Using Evolutionary frameworks to Study Epigenetic Aging – the Epigenetic PaceMaker In multiple studies DNA methylation has proven to be an accurate biomarker of age. To develop these biomarkers, the methylation of multiple CpG sites is typically linearly combined to predict chronological age [1]. By contrast, in this study we apply the Universal PaceMaker (UPM) model to investigate changes in DNA methylation during aging. The UPM was initially developed by us in a separate project to study rate acceleration/deceleration during genome evolution. Here, rather than identifying which linear combinations of sites predicts age as in the traditional approach, the new framework, the Epigenetic PaceMaker (EPM) models the rates of change
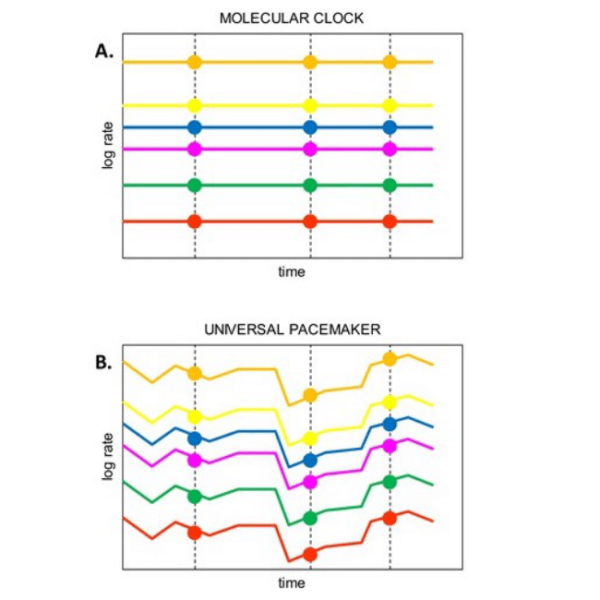
Universal PaceMaker of Genome Evolution
Universal PaceMaker of Genome Evolution Here we are collaborating with the Evolutionary Genomics group of Eugene Koonin at the NIH. We were awarded the BSF grant on a project tackling prokaryotic evolution, but are now looking at another, somehow broader, more fundamental evolutionary phenomenon. This project concerns with correlation between evolutionary rates of the various genes in the genome and was originated from our other project dealing with identifying HGT. Gene evolution is traditionally considered within the classical framework of the Molecular Clock (MC) model [1] according to which each gene is characterized by an approximately constant rate of evolution. However, in a recent comparative analysis of numerous gene histories (gene phylogenies)
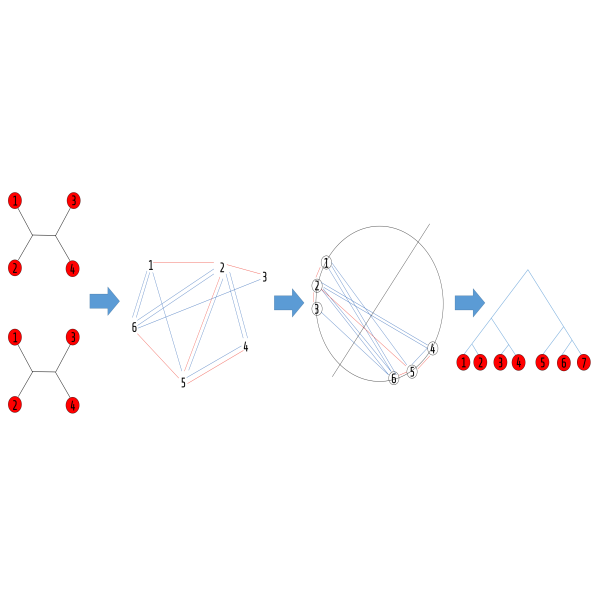
MaxCut Supertree
MaxCut Supertree A quartet tree (or simply a quartet) is a tree over four taxa (species) that is the most basic phylogenetic building block – A tree can be decomposed into its defining quartets that uniquely define it. It is this fundamentality that confers the quartets their power. The converse however, is not true – it is rare that an arbitrary quartet set agrees on some tree. Hence, given a set of quartets, it is a common practice to look for a tree agreeing with the most of them. This seemingly simple and “clean” question lies at the heart of almost any specific, more complicated question in phylogenetics. Due to
Convex Recoloring of Trees and Networks
Convex Recoloring of Trees and Networks Definition: Given a necklace of colored beads, we say that the string is convex if all beads of the same color are in one chunk. In graph theory terms, we can view the necklace as a path and the coloring C as function from V to the set of colors C . Then C is convex if every subset of vertices belonging to the same color class is connected . However, when the coloring is not convex, we would like to find the closest coloring C’ that is convex. This distance between the two colorings C’ and C depends on the model assumed. Under the simplest model this is just the number of vertices whose color is changed between C’ and C . It can be shown that under this model,
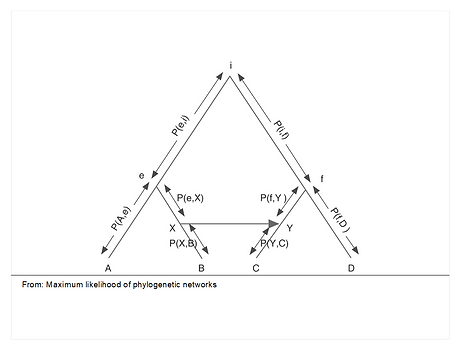
Statistical Analysis of Trees and Networks
Statistical Analysis of Phylogenetic Trees and Networks The study of Evolution and understanding the history of life on Earth, has attracted scientists’ attention since the dawn of modern science. Phylogenetics, the reconstruction of this history, has expanded beyond its traditional role in evolutionary studies and with the advent of next generation sequencing is increasingly integrated into modern biological areas such as preventive medicine, epidemiology, and human migration. Maximum Likelihood (ML) is considered as the most accurate phylogenetic method. Our lab has developed expertise in analytical solutions to ML phylogenetic reconstruction. The novelty of our approach is the representation of the ML problem as a constraint optimization problem and using algebraic

